Press Releases
* Please note that the news release contains the content at the time of the announcement and may differ from the latest information.
Succeeded in highly efficient genome editing in Euglena
-Expectations for useful Euglena breeding for biofuel production-
Euglena Co., Ltd.
RIKEN
Toshinao Nomura, Team Leader, Bioproduction Information Research Team, RIKEN (RIKEN), Center for Environmental Resource Science, Kengo Suzuki, Team Leader, Microalgae Production Control Technology Research Team, Baton Zone Research Promotion Program of the Science and Technology Hub Headquarters (Co., Ltd.EuglenaExecutive Officer R&D / Director of Advanced Technology Research) and other research teams are working on the industrial use species of the midges. Euglena gracilis[1] High-efficiency genome editing for (hereinafter referred to as "green beetle") [2] For the first time we succeeded in establishing a method.
The results of this research are expected to greatly contribute to the promotion of basic research on green beetles and the breeding of useful strains.
Astragalus is a useful microalgae whose application to food and biofuels is underway. However, an efficient genome editing method that is important for advancing basic research and useful strain breeding has not been established until now.
In this study, the research team found that the Cas9 RNP complex was used in Astragalus cells. [3] By using the direct introduction method and finding the optimal conditions, we succeeded in introducing defects and insertions and mutations into the target gene with a very high editing efficiency of about 80%. Furthermore, it was clarified that it is possible to introduce long defective mutations into specific regions of DNA using two types of Cas9 RNP complexes, and to introduce accurate foreign DNA molecules using single-stranded donor DNA molecules.
The study was published in the British scientific journal Plant Biotechnology Journal was published in an online version (dated May 26) prior to publication.
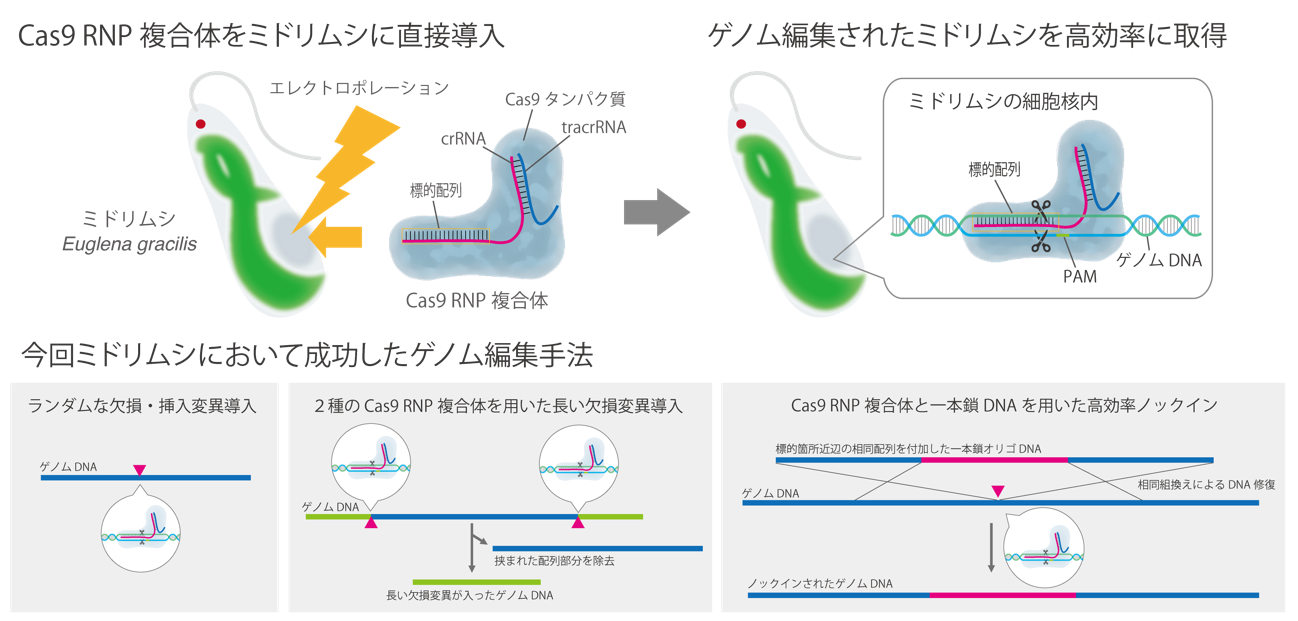
Figure Outline of genome editing method using Cas9 RNP in Euglena that succeeded this time
* Research team
RIKEN
Bio-Production Information Research Team, RIKEN Center for Sustainable Resource Science
Researcher Toshihisa Nomura (Toshihisa Nomura)
((RIKEN Science and Technology Hub Industrial Federation Headquarters Baton Zone Research Promotion Program Microalgae Production Control Technology Research Team Researcher)
Technical Staff II Komaki Inoue
Technical Staff II Uehara (Yamaguchi) Yukiko (Uehara (Yamaguchi) Yukiko)
Team leader Keiichi Mochida (Keiichi Mochida)
((RIKEN Science and Technology Hub Industrial Federation Headquarters Baton Zone Research Promotion Program Microalgae Production Control Technology Research Team Deputy Team Leader)
Science and Technology Hub Industry Federation Headquarters Baton Zone Research Promotion Program
Microalgae Production Control Technology Research Team
Visiting Researcher Yasushi Yamada (Koji Yamada)
(Team Leader, Advanced Technology Research Division, Advanced Technology Research Department, Euglena Co., Ltd.)
Visiting Researcher Osamu Iwata (Osamu Iwata)
(Euglena Co., Ltd. Corporate Strategy Department)
Team leader Kengo Suzuki
(Euglena Co., Ltd. Executive Officer in charge of R & D / General Manager of Advanced Technology Research Department)
1. 1. background
In recent years, the realization of a bioeconomy that balances sustainability and economic activity, and the Sustainable Development Goals (SDGs) [Four] It is expected to develop technology for producing useful substances such as biofuels and bioplastics using microalgae in order to achieve the above.
Among them, the industrially used species of Euglena, for which mass culture methods have been established indoors and outdoors. Euglena gracilis (Euglena) is a stored polysaccharide paramylon that can be used as a bioplastic material and functional food under aerobic conditions. [Five] Accumulates (Fig. 1). On the other hand, under anaerobic conditions, paramylon produces fats and oils called wax esters. Wax esters are suitable for jet fuel and are currently being tested for practical use. Euglena is also highly nutritious and is attracting attention as a feed for which demand is expanding worldwide. Against this background, it is required to accumulate biological knowledge for utilizing Euglena and to create modified strains with useful traits.
However, a practical and sustainable method for modifying the gene function of Euglena has not yet been established, which has hindered basic research and efficient breeding. Therefore, the research team started research with the aim of establishing a genome editing method suitable for practical use, considering the possibility of future industrial use.
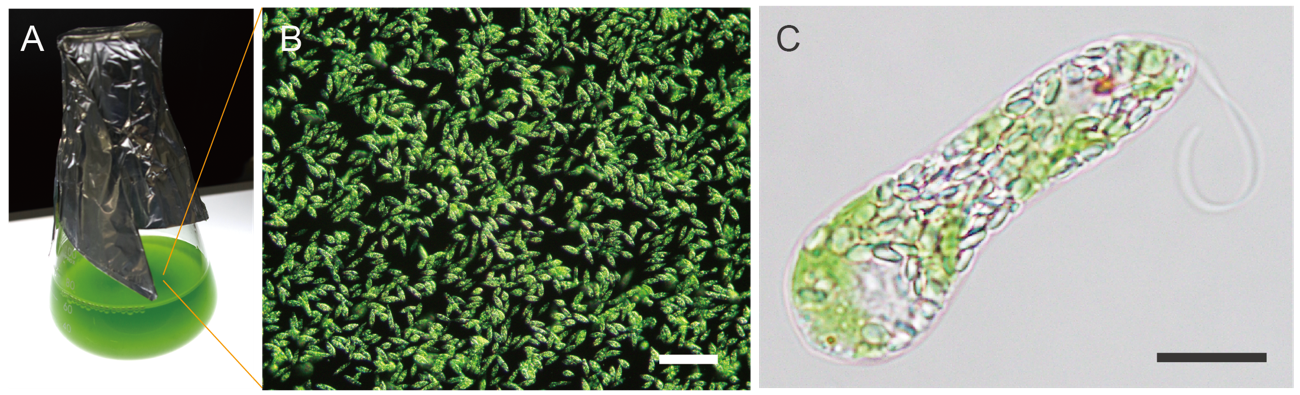
Fig. 1 Euglena gracilis culture under aerobic conditions and micrographs
(A) Euglena cultured in a laboratory with shaking using a liquid medium.
(B) Euglena growing in the culture medium. The scale bar is 100 micrometers (μm, 1/1000 mm).
(C) Microscopic image of one Euglena. Paramylon granules are a large number of small particles found in cells. The scale bar is 10 μm.
2. 2. Research methods and results
First, the research team started with genes for enzymes involved in the synthesis of paramylon in Euglena. EgGSL2[6] Guide RNA (gRNA) targeting [3] We designed a Cas9 RNP complex with cleaving activity for the target DNA. Next, morphological changes of paramylon grains and T7E1 assay method [7] Electroporation method using [8] We searched for the optimum conditions for directly introducing the Cas9 RNP complex into Euglena cells (Figs. 2 and 3).
As a result, amplicon sequence analysis [9] In the evaluation by, we succeeded in introducing a random deletion / insertion mutation with a high efficiency of about 80% 72 hours after the introduction of the Cas9 RNP complex, which is unprecedented in other microalgae.
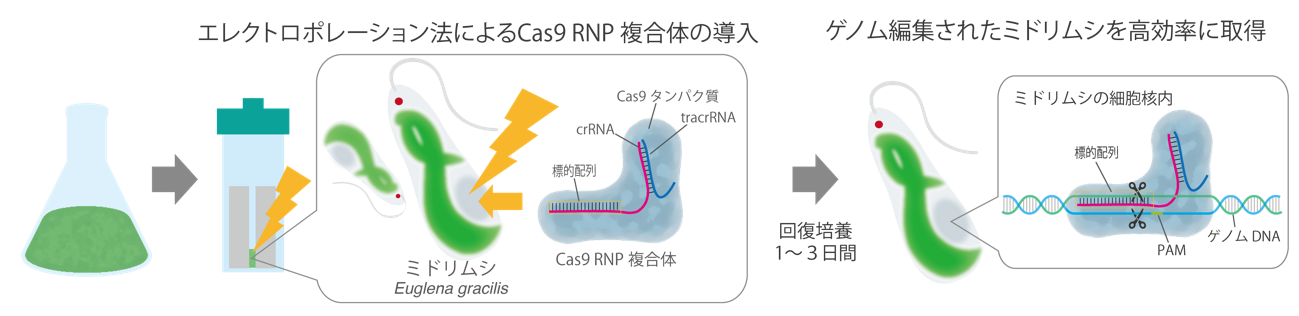
Fig. 2 Flow of Euglena gracilis genome editing method using Cas9 RNP complex
First, gRNAs (crRNA and tracrRNA) corresponding to the target gene EgGSL2 of Euglena are designed to prepare a Cas9 RNP complex. This is directly introduced into Euglena by the electroporation method. At this time, by using the optimized conditions, it became possible to acquire the genome-edited individual with high efficiency and in a short time.
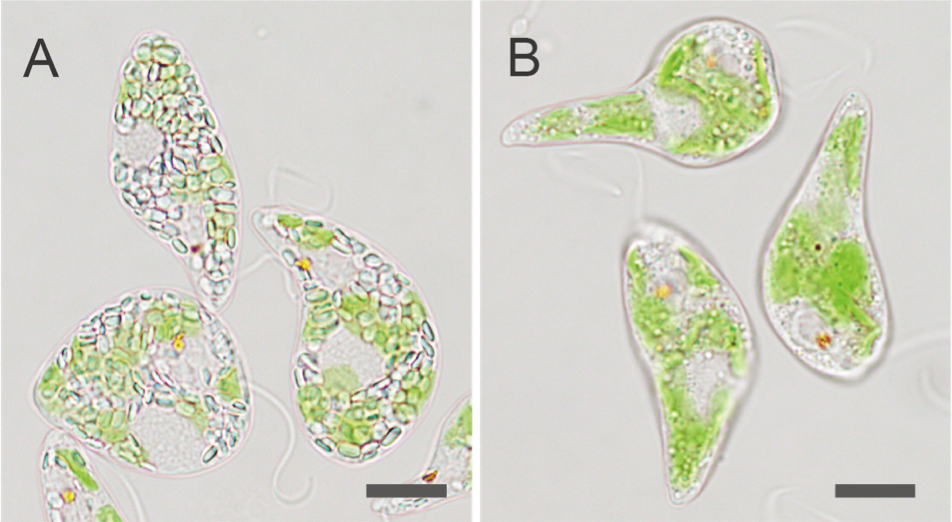
Fig. 3 Example of Euglena gracilis produced by genome editing using Cas9 RNP complex
An example of a genome editing strain created by introducing (A) a wild strain of Euglena and (B) a Cas9 RNP complex that targets the EgGSL2 gene of an enzyme involved in paramylon synthesis. In Euglena B, the function of the EgGSL2 gene is lost, so that the accumulation of paramylon grains generated under heterotrophic culture is hardly observed in the wild strain. The scale bar is 10 μm.
In addition, the simultaneous introduction of two types of Cas9 RNP targeting distant locations on the genome into Euglena cells was able to induce long deletion mutations. Furthermore, ssODN (single-stranded oligodeoxynucleotide) [10] with sequences homologous to the upstream and downstream of the target sequence site added to both ends together with the Cas9 RNP complex should be introduced into Euglena cells as donor DNA. We also succeeded in an accurate small-scale (short base sequence) knock-in [11].
3. 3. Expectations for the future
In this study, we succeeded in establishing a DNA-free genome editing method for euglena with a high efficiency that is unparalleled in other microalgae. Furthermore, based on the highly efficient genome editing method described above, we have also succeeded in efficient knock-in experiments using ssODN, making it possible to modify the Euglena genome in a more precisely controlled manner.
In the future, by making full use of this technology, in addition to promoting basic research such as the analysis of gene functions in euglena, it will be possible to develop molecular breeding strains with improved ability to produce useful substances and to create euglena designed according to the purpose. You can expect it to be.
In addition, of the 17 "Sustainable Development Goals (SDGs)" set by the United Nations in 2016, this research is based on "7. Affordable and clean energy" and "13. Concrete measures against Climate Change." This achievement is expected to contribute to
4. Treatise information
<Title>
Highly efficient transgene-free targeted mutagenesis and single-stranded oligodeoxynucleotide-mediated precise knock-in in the industrial microalga Euglena gracilis using Cas9 ribonucleoproteins
<Author name>
Toshihisa Nomura, Komaki Inoue, Yukiko Uehara-Yamaguchi, Koji Yamada, Osamu Iwata, Kengo Suzuki, Keiichi Mochida
<Magazine>
Plant Biotechnology Journal
<DOI>
10.1111 / pbi.13174
5. Supplementary explanation
[1] Euglena gracilis
It is a kind of microalgae of the genus Euglena that grows in rice fields and freshwater lakes, and has been used for biological experiments since ancient times. Since the mass culture method has been established, this species is most suitable for industrial use among Euglena, and its utilization for various purposes is being developed.
[2] Genome editing
A technique for modifying genetic information by allowing a nucleic acid-degrading enzyme (nuclease) or the like to act site-specifically.
[3] Cas9 RNP complex, guide RNA (gRNA)
The Cas9 RNP complex is a stable ribonuclear protein complex composed of gRNA and Cas9 protein designed based on the target site on the genome, and specifically cleaves the target DNA site on the genome corresponding to the gRNA. .. GRNA in genome editing refers to crRNA (CRISPR RNA) and tracrRNA (trans-activating crRNA) or single guide RNA. Cas9 RNP is an abbreviation for Cas9 (CRISPR associated protein 9) / gRNA (guide RNA) Ribonucleoproteins.
[4] Sustainable Development Goals (SDGs)
International goals from 2016 to 2030 set out in the 2030 Agenda for Sustainable Development adopted at the United Nations Summit in September 2015. Consisting of 17 goals and 169 targets for realizing a sustainable world, it is a universal thing that not only developing countries but also developed countries themselves work on, and Japan is also actively working on it. (Reprinted from the Ministry of Foreign Affairs website with some modifications)
[5] Paramylon
A storage polysaccharide in Euglena to which glucose (dextrose) is β-1,3 bound. When accumulated intracellularly in Euglena, it is seen as a granular structure. In addition, under anaerobic conditions, fats and oils (wax esters) are produced based on paramylon in order to obtain energy.
[6] EgGSL2 gene
An enzyme gene involved in the synthesis of paramylon in Euglena. Inhibition of this gene reduces paramylon accumulation under heterotrophic culture conditions. EgGSL2 is an abbreviation for E. gracilis Glucan Synthase-Like 2.
[7] T7EI assay
A method that uses T7 Endonuclease I (T7EI), an enzyme that recognizes and cleaves DNA mismatches, to easily detect the presence or absence of deletion / insertion mutations due to genome editing.
[8] Electroporation method
Also called electroporation, a method in which a minute hole is temporarily made in the cell membrane by an electric pulse to directly introduce a substance (Cas9 RNP complex and ssODN in this study).
[9] Amplicons sequence analysis
An analysis method that amplifies specific sites on the genome (target sites in the EgGSL2 gene in this study) and comprehensively acquires their nucleotide sequence information using a next-generation sequencer.
[10] ssODN (single-strand oligodeoxynucleotide)
Single-stranded DNA. By introducing ssODN as a donor DNA into cells together with an RNP complex or the like, knock-in or base substitution to the genome based on the homology repair mechanism becomes possible.
[11] Knock-in
A genetic engineering technique that inserts DNA into a target site on the genome.
6. Presenter / institution window
<Presenter> * Please contact the presenter for research content.
RIKEN
RIKEN Center for Sustainable Resource Science
Bio-Production Information Research Team
Researcher Toshihisa Nomura (Toshihisa Nomura)
Team leader Keiichi Mochida (Keiichi Mochida)
Science and Technology Hub Industry Federation Headquarters Baton Zone Research Promotion Program
Microalgae Production Control Technology Research Team
Team leader Kengo Suzuki
(Euglena Co., Ltd. Executive Officer in charge of R & D / General Manager of Advanced Technology Research Department)
TEL:045-503-7077(野村、持田) FAX:045-503-9609(野村、持田)
E-mail:toshihisa.nomura[at]riken.jp(野村), keiichi.mochida[at]riken.jp(持田)

-Contact for inquiries from the press-
Euglena Co., Ltd. Corporate Communication Division
RIKEN Public Relations Office Press
